Machine data analysis using artificial intelligence
A Generic Pipeline for AI-based Data Analysis
by DI Dr. Alexander Maletzky
Nowadays, data is recorded and stored in vast quantities. The goal is often to create a data-based forecasting model that can be used to predict future developments. However, the path from the raw data to the finished model is usually longer than expected. RISC Software GmbH developed a generic pipeline for AI-based data analysis.
Table of contents
- The problem
- Our solution: A generic pipeline
- Example application: Mortality prediction in the intensive care unit
- Further information
- Author

The problem
State-of-the-Art Methoden der künstlichen Intelligenz, wie z.B. neuronale Netze, benötigen qualitativ hochwertige, gut aufbereitete Daten als Input, um brauchbare Ergebnisse erzielen zu können. Die Realität hält diesen Anforderungen jedoch nicht Stand: Daten enthalten Ausreißer und fehlende Werte, oder werden in unterschiedlichen – manchmal sogar unregelmäßigen – Messfrequenzen aufgezeichnet.
What to do?
In order to be able to use the data nevertheless, extensive preparation is necessary. Of course, this process depends on the specific data, but essentially always involves the same steps:
- Importing the raw data, e.g. from relational databases,
- Validating and harmonising the data, and
- Importing (“filling in”) missing values.
For the downstream training of a predictive model, “organisational” steps are also necessary, such as partitioning into training and test data. Figure 1 schematically depicts the entire process of machine data analysis.

Our solution: A generic pipeline
As part of the MC3 project (https://risc-software.at/mc3/), which deals with data analysis in the medical environment, experts from RISC Software GmbH have developed a generic data pipeline. This allows a large part of the data analysis process to be mapped – in particular, the aforementioned data preparation is an integral part. In addition, the system provides a uniform interface for any machine learning algorithms or model classes, so that the training, application and analysis of a prediction model can also be mapped via the pipeline.
A special focus is also on the increasingly important topic of Explainable AI – explainable artificial intelligence. Thus, almost any state-of-the-art explanation method, from Layerwise Relevance Propagation to Shapley Values, can be integrated via a simple interface to make model predictions comprehensible to humans. The pipeline is implemented in such a way that it is as reusable as possible. It is modular, which means that individual components can be combined, added and removed as desired. In addition, end users can easily configure the individual steps, such as specifying validation rules, imputation strategies, etc. Even grid search for exploring the parameter space is easily possible. The applicability of the pipeline is thus not limited to medical data.
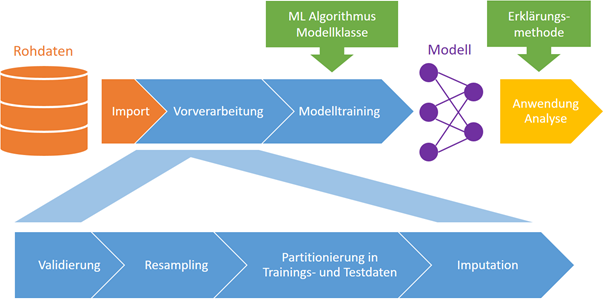
Figure 1. Schematic representation of machine data analysis using artificial intelligence. The reusable data pipeline includes the components shown in blue, and supports data import and application/analysis of the trained model.
Example application: Mortality prediction in the intensive care unit
Researchers at RISC Software GmbH applied the pipeline as an example to the publicly available MIMIC-III database, which is often used in the literature as a benchmark dataset in the field of (intensive) medical data analysis. The aim was to predict the probability of death of a patient in intensive care based on the first twelve hours after admission. Thanks to the pipeline developed, almost the entire data analysis and modelling process could be reduced to a few simple parameter configurations. The result achieved does not have to fear comparison with current scientific publications on this topic.
The projects of the Medical Informatics Department are funded by the Upper Austrian government under the strategic economic and research programme “Innovative Upper Austria 2020”.
Further information
Technology stack: Python 3, with the relevant add-on packages (Pandas, Plotly, scikit-learn, etc.).
MC³: Medical Cognitive Computing Center, joint research project of Kepler University Hospital Linz / MedCampus III, Johannes Kepler University Linz / Institute for Machine Learning, and RISC Software GmbH / Department of Medical Informatics.
MIMIC-III database: Medical Information Mart for Intensive Care III, public dataset of over 58,000 intensive care patients from a hospital in Boston, MA; https://mimic.physionet.org/
Contact
Author

DI Dr. Alexander Maletzky
Researcher & Developer Unit Medical Informatics